PLEK is a novel alignment-free tool.
10-fold cross-validation on the human training datasets
was performed. The accuracy of PLEK was 95.6%.
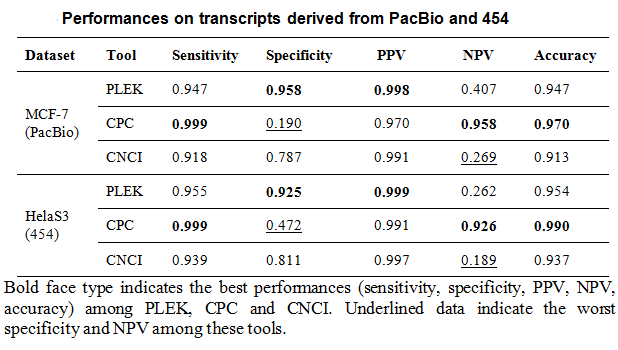
PLEK achieved an optimal balance between
high specificity and high sensitivity (0.946 and 0.942 on
PacBio, 0.955 and 0.925 on 454).
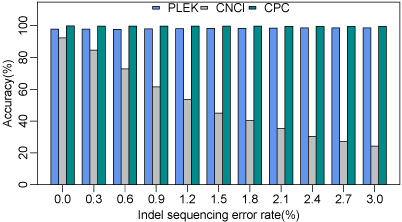
Comparison of robustness towards indel sequencing errors. The x-axis is the
indel numbers per 100 bases (indel sequencing error rates). Performance (accuracy) of CNCI
declines significantly as the indel error rate increases.
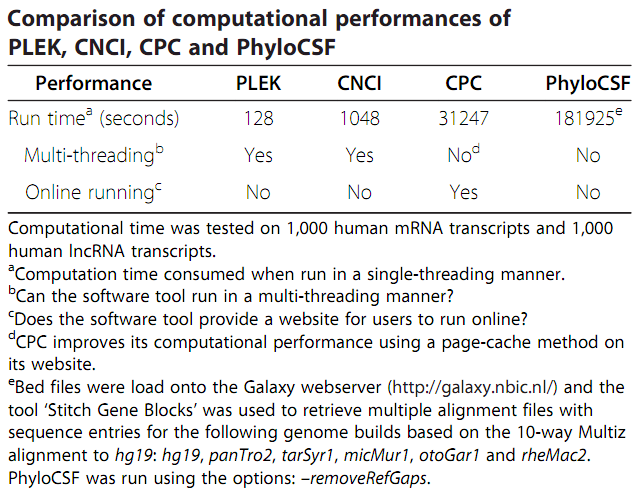
Several tools was tested on 1,000 human mRNA transcripts and 1,000 human lncRNA transcripts. PLEK took 128 seconds to process the data, which was approximately
eightfold faster than CNCI (1,048 seconds), 244 times than CPC (31,247 seconds) and 1,421
times than PhyloCSF (181,925 seconds). Additionally, PLEK could be easily
configured for multi-threading parallel computing, which will further save computation time.
Thus, PLEK is especially suitable for classifying a large number of transcripts conducted by
RNA-seq technologies.