Help
Target
Figure A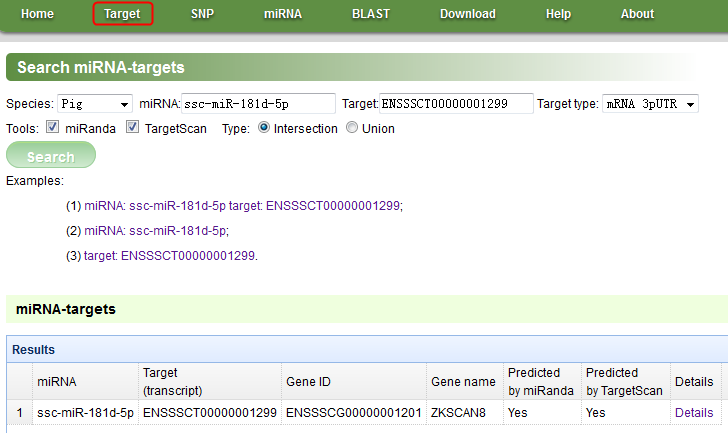
NOTE: In this page, we can search/view miRNA-targets (miRNA-mRNA and miRNA-lncRNA interactions). Click the ‘Target’ menu in the navigation bar to open this page, and click the examples to learn about how to use.
Species: currently, we can choose from ‘Pig’, ‘Chicken’ and ‘Cow’.
miRNA: miRNA ID.
Target: It can be an Ensembl mRNA transcript ID, gene name, gene ID or an ALDB lncRNA ID.
Target type: ‘mRNA 3pUTR’, search binding sites in 3’UTR of mRNAs; ‘mRNA CDS’, search binding sites in CDS of mRNAs; ‘mRNA 5pUTR’, search binding sites in 5’UTR of mRNAs; ‘lncRNA, search binding sites in lncRNAs.
Tools: which tools were used to predict targets? miRanda and/or TargetScan.
Type: ‘Intersection’, only present the binding interactions that are predicted by both miRanda and TargetScan. ‘Union’, present the binding interactionss that are predicted by miRanda and/or TargetScan.
Examples: there are three examples. Click one of their links to see an example.
Figure B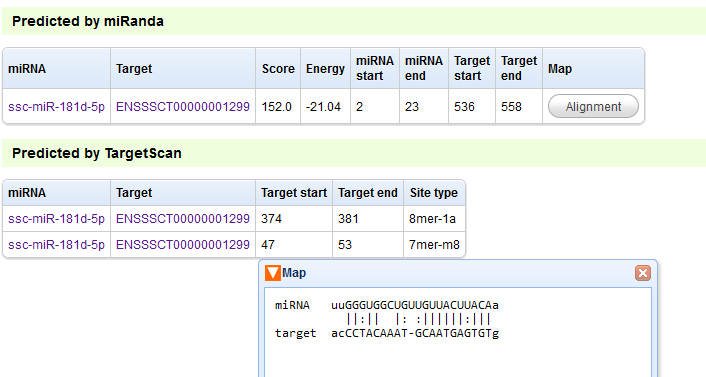
NOTE: In this page, we can view the details of interactions and visualize the binding maps. Click the ‘Details’ link to open this page. Click ‘Alignment’ to visualize the ‘Map’ relation.
miRNA: miRNA ID. Click the link to see details of the miRNA.
Target: Ensembl mRNA ID or ALDB lncRNA ID. Click the link to see details of the RNA.
Score: miRanda score.
Energy: miRanda minimal free energy.
miRNA start: the start position of miRNA that binds to target.
miRNA end: the end position of miRNA that binds to target.
Target start: the start position of target that binds to miRNA.
Target end: the end position of target that binds to miRNA.
Map: visualize the binding of miRNA to target.
Site type: site type of TargetScan.
SNP
Figure C
NOTE: In this page, we can search/view the interactions which are predicted by both miRanda and TargetScan. We also can investigate whether there are SNPs in overlapping binding sites or miRNAs. Click the ‘SNP’ menu in the navigation bar to open this page, and click the first example to see an example.
Species: currently, we can choose from ‘Pig’, ‘Chicken’ and ‘Cow’.
miRNA: miRNA ID.
Target: It can be an Ensembl mRNA transcript ID, gene name, gene ID or an ALDB lncRNA ID.
Target type: ‘mRNA 3pUTR’, search binding sites in 3’UTR of mRNAs; ‘mRNA CDS’, search binding sites in CDS of mRNAs; ‘mRNA 5pUTR’, search binding sites in 5’UTR of mRNAs; ‘lncRNA, search binding sites in lncRNAs.
Type: ‘ No overlapping SNP’, there are no SNPs in miRNAs or binding sites of targets; ‘SNP in miRNA’, there are SNPs in miRNAs; ‘SNP in binding sites’, there are SNPs in overlapping binding sites of targets; ‘SNP in miRNA and binding sites’, there are SNPs in both miRNAs and overlapping binding sites; ‘All’, all results.
Examples: there are three examples. Click one of their links to see an example.
Overlapping sites: the overlapping binding sites which are predicted by both miRanda and TargetScan. Format: ‘chromosome|strand|start position|end position’.
Target-SNP chromosome: the chromosome of SNPs which are located in overlapping binding sites.
Target-SNP location: the location of SNPs which are located in overlapping binding sites.
miRNA-SNP chromosome: the chromosome of SNPs which are located miRNAs.
miRNA-SNP location: the location of SNPs which are located in miRNAs.
miRNA
Figure D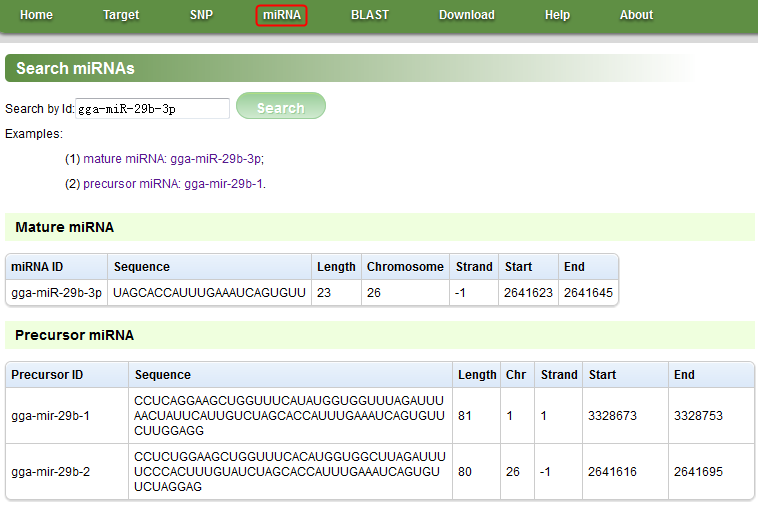
NOTE: In this page, we can search/view mature and precursor miRNAs. Click the ‘miRNA’ menu in the navigation bar to open this page, and click the first example to see an example.
Search by Id: Mature miRNA ID or precursor miRNA ID.
Examples: there are two examples. Click one of their links to see an example.
miRNA ID: miRNA ID.
Sequence: Sequence of mature/precursor miRNA.
Length: Length of mature/precursor miRNA.
Chromosome: Chromosome of mature miRNA.
Strand: Strand of mature/precursor miRNA.
Start: Start of mature/precursor miRNA.
End: End of mature/precursor miRNA.
Precursor ID: Precursor ID.
Chr: Chromosome of precursor miRNA.
BLAST
Figure E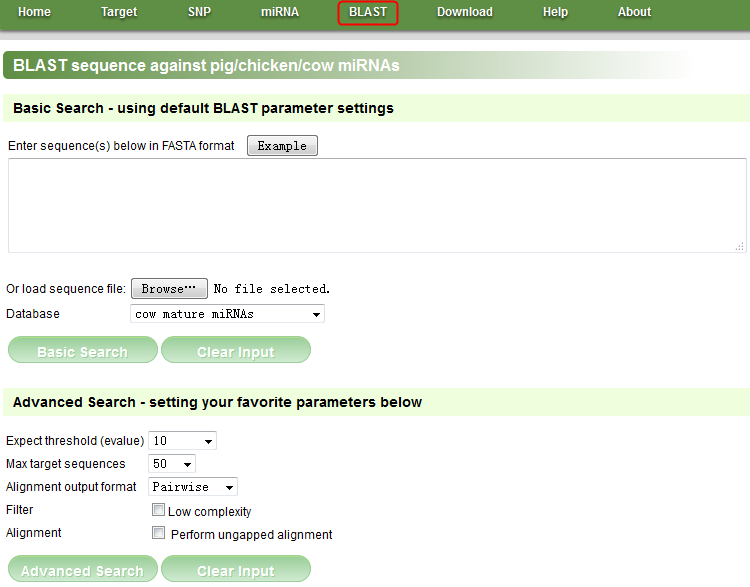
NOTE: In this page, we can blast sequences to pig/chicken/cow mature and precursor miRNAs. Click the ‘BLAST’ menu in the navigation bar to open this page, and click the ‘Example’ button, then click the ‘Basic Search’ button to see an example.
Download
Figure F
NOTE: In this page, we can download predicted/processed results for further study. Click the ‘Download’ menu in the navigation bar to open this page.
