Help
Nomenclature of genes and transcripts in RatTransc
Format: (RatTransc|ENS)(RNO)(G|T)##########RatTransc is a prefix. The genes or transcripts with this prefix are annotated by our approach.
ENS is a prefix. The genes or transcripts with this prefix are annotated by ENSEMBL.
RNO - Rattus norvegicus.
G|T G - gene; T - transcript.
########## a serial number.
e.g. RatTranscG003693 is the ID of a Rattus norvegicus gene; RatTranscT010140 is the ID of a Rattus norvegicus transcript.
Search
A search function was designed to facilitate users to retrieve useful information, such as the basic information of transcripts and genes, expression profiles, neighboring genes, overlap with QTLs and the lncRNA genes adjacent to a specific protein-coding gene. These functional units are contained in the “Search” section on the main navigation bar. This section mainly includes “Search by Id/Name”, “Search by Genomic Location”, and “Search Neighboring LncRNAs” pages.
In the “Search by Id/Name” page, user can find transcripts and genes by inputting their ids(type lincRNA's ID defined by this project or protein-coding's Ensembl ID) or names (Figure A). For a transcript/gene, the following relative information will be represented: “Basic information” (Figures B, C), “Expression profile” (Figure D), “Neighboring genes” (Figure E). The content panels can be expanded or collapsed when their head titles are clicked.
The “Search by Genomic Location” page is designed to help users to retrieve all the genes and transcripts located in a specific genomic region (Figure F). If users are interested in a transcript or gene, they can click its id in the first column to view its details.
The “Search Neighboring LncRNAs” page can be used to search the lncRNA genes adjacent to a protein-coding gene (Figure G).
Figure A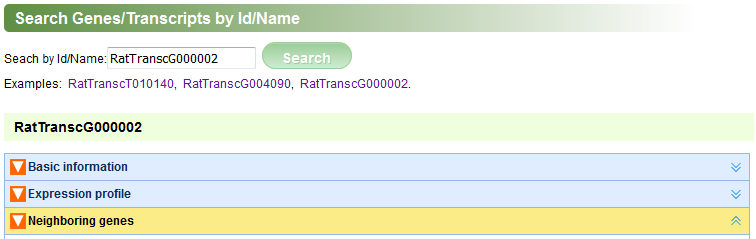
Figure B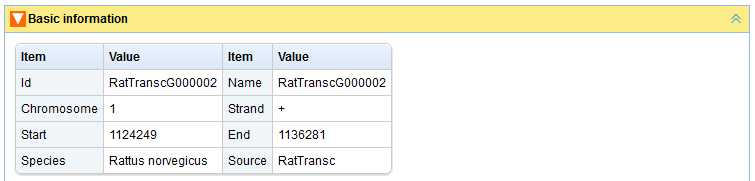
Figure C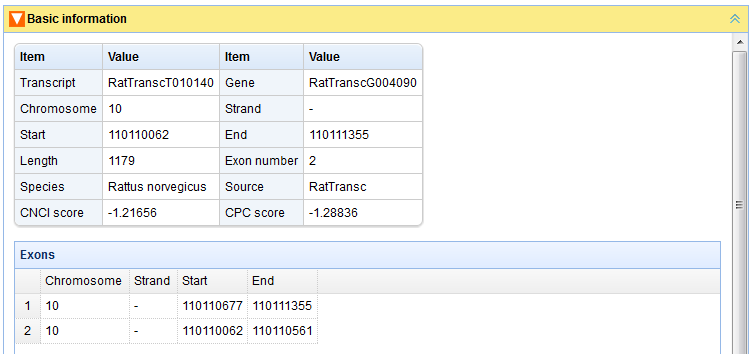
Figure D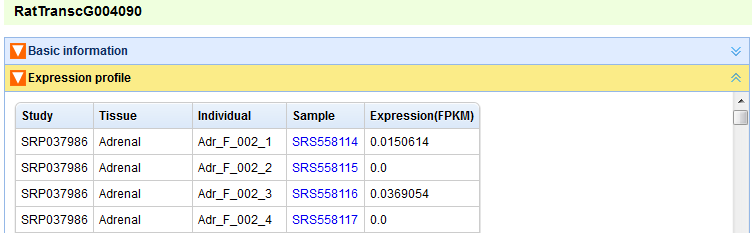
Figure E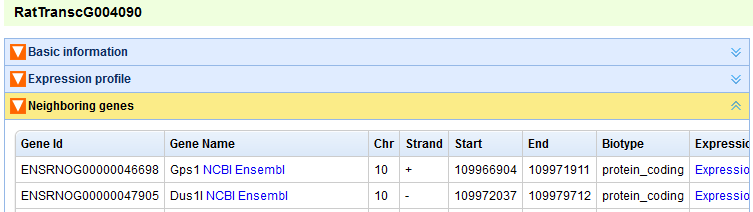
Figure F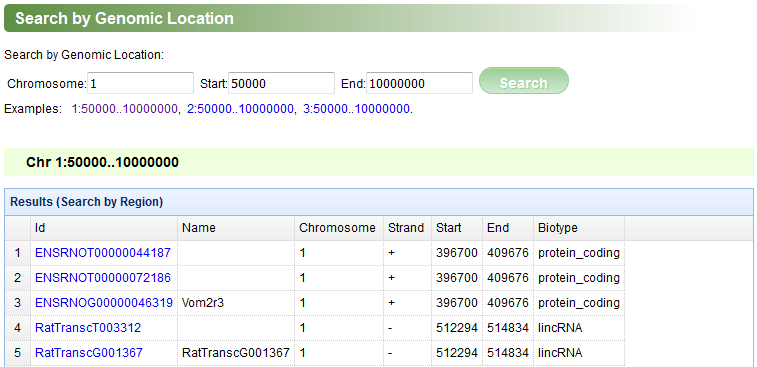
Figure G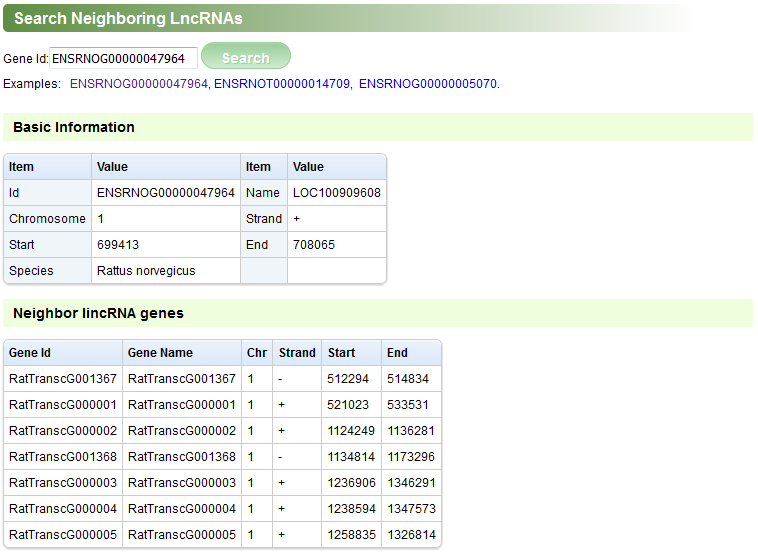
BLAST
Since RatTransc maintains nucleotide sequence data of lncRNAs, a sequence alignment tool, called wwwblast, based on the Basic Local Alignment Search Tool (BLAST) is integrated into RatTransc. Users can compare a query sequence with a library of lncRNA sequences using a BLAST search, and identify lncRNA sequences that closely resemble the query sequence. BLAST will meet users’ requirements for various interests related to homologous transcripts or genes. On the BLAST page, users can supply one or more query sequences by uploading or directly pasting them to search against the available databases using BLAST's default parameters. Users can also specify more parameters for BLAST search to control the search sensitivity, the result format, etc. The BLAST results will be displayed in another page in the Pairwise format by default.
Analysis
Tissue-specific Expression This page lists the tissue-specific expression genes. For each study, we take
one tissue as CASE tissue, and the rest as CONTROL tissue. The results were derived by using DESeq2 and were
filtered by padj<0.1 (padj<0.1 remained).
Blast lncRNAs among Species We blasted lncRNA transcript sequences among human and mouse.
Download
Data from the RatTransc can be downloaded to perform local analysis. In the Download page, users can download lncRNAs in FASTA, GFF3 and GTF formats. They are divided per species and per content type, with information provided for the assembly of the species . We also provide the expression levels which we derived from the datasets SRP037986 and SRP041119. The FPKM (fragments per million reads per kb of transcript) expression levels were obtained by TopHat (with the parameter --no-novel-juncs) and Cufflinks (with the parameter -G). The count expression levels were generated by summaryOverlaps with the parameter mode="Union".
NOTE: Some files were compressed using tar. We can uncompress them with tar. For example, tar -zxvf RatTransc_lincRNAs.v1.fa.tar.gz
